A team of researchers at the University of Washington collaborated to address challenges in protein sequence design methods using LigandMPNN, a deep learning-based protein sequence design method. This model targets the design of enzymes, small molecule binders, and sensors. Existing physics-based approaches such as Rosetta and deep learning-based models such as ProteinMPNN cannot explicitly model non-protein atoms and molecules, and this limitation makes it possible to interact with small molecules, nucleotides, and metals. Precise design of acting protein sequences has been hampered.
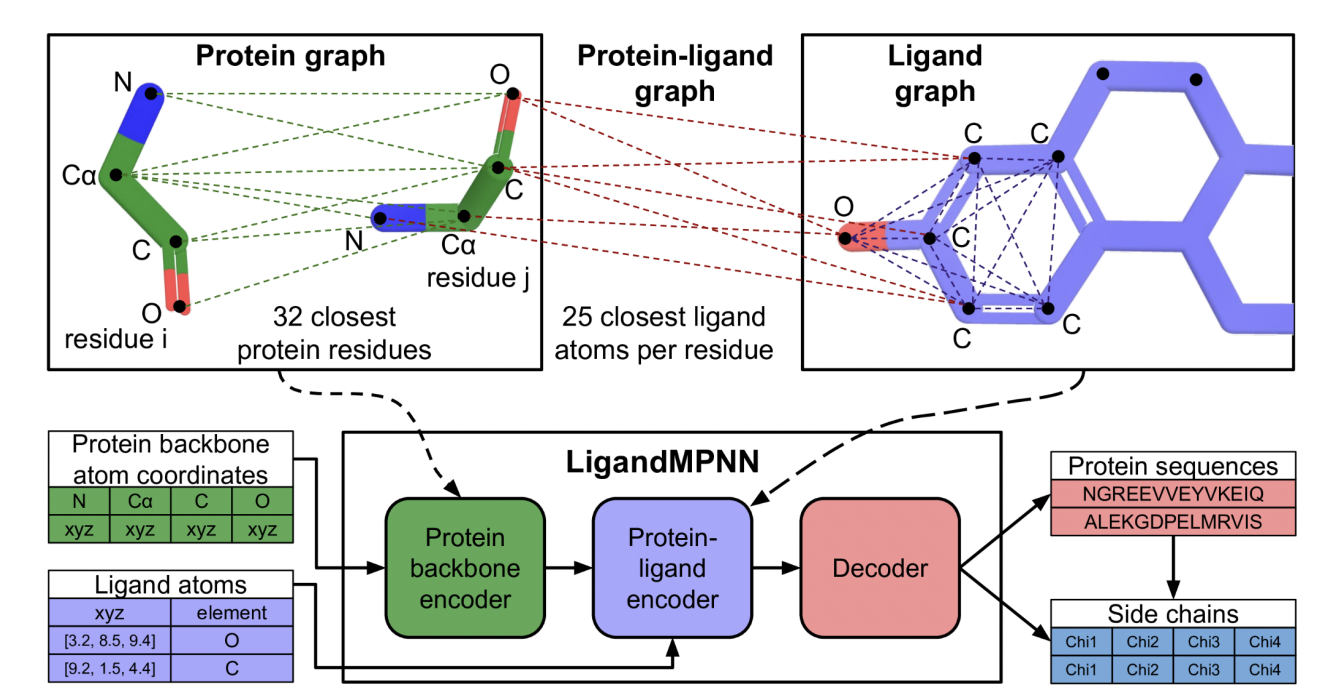
The aforementioned methods ignore explicit consideration of non-protein atoms and molecules, which are important for the design of enzymes, protein-DNA/RNA interactions, protein-small molecules, and protein-metal binders. The proposed solution, LigandMPNN, is built on the ProteinMPNN architecture but explicitly incorporates a complete non-protein atomic context. LigandMPNN leverages neural networks to model interactions and introduces a protein-ligand graph that encodes the shape of the ligand atoms. This modification allows LigandMPNN to generate sequences and side chain conformations tailored to specific non-protein contexts.
LigandMPNN adopts a graph-based approach, treating protein residues as nodes and incorporating the nearest neighboring edges based on the Cα-Cα distance. This model captures interactions by introducing a protein-ligand graph that represents the geometric relationships between protein residues and ligand atoms as nodes and edges. Ligand graphs enhance the communication of information to proteins through ligand and protein edges.
This experiment demonstrates the superior performance of LigandMPNN and its side chain packing compared to Rosetta and ProteinMPNN, with 20-30% higher sequence recoveries for residues that interact with small molecules, nucleotides, and metals. The accuracy was improved and its effectiveness in detailed structural design was demonstrated. LigandMPNN also outperforms existing models in terms of speed and efficiency. LigandMPNN is approximately 250 times faster than Rosetta.
In conclusion, LigandMPNN fills a critical gap in existing protein sequence design methods by explicitly including non-protein atoms and molecules. LigandMPNN’s graph-based approach shows significant improvements in performance, leading to higher sequence recoveries and superior side chain packing accuracy around small molecules, nucleotides, and metals. LigandMPNN exhibits excellent performance in designing small molecules and DNA-binding proteins with high affinity and specificity, greatly aiding protein engineering.
Please check paper. All credit for this study goes to the researchers of this project.Don’t forget to follow us twitter and google news.participate 36,000+ ML SubReddits, 41,000+ Facebook communities, Discord channeland linkedin groupsHmm.
If you like what we do, you’ll love Newsletter..
Don’t forget to join us telegram channel
Pragati Jhunjhunwala is a consulting intern at MarktechPost. She is currently pursuing her bachelor’s degree from Indian Institute of Technology (IIT), Kharagpur. She is a technology enthusiast and has a keen interest in software and data and a range of science applications. She is constantly reading about developments in various areas of AI and ML.
 Join the fastest growing AI research newsletter from researchers at Google + NVIDIA + Meta + Stanford + MIT + Microsoft and more…
Join the fastest growing AI research newsletter from researchers at Google + NVIDIA + Meta + Stanford + MIT + Microsoft and more…